Within the twentieth century, when a regimen an infection was once handled with an ordinary antibiotic, restoration was once anticipated. However through the years, the microbes accountable for those infections have advanced to evade the very medicine designed to get rid of them.
Each and every yr, there are greater than 2.8 million antibiotic-resistant infections in the US, resulting in over 35,000 deaths and US$4.6 billion in well being care prices. As antibiotics grow to be much less efficient, antimicrobial resistance poses an expanding danger to public well being.
Antimicrobial resistance started to emerge as a significant danger within the Forties with the upward push of penicillin resistance. By means of the Nineteen Nineties, it had escalated into an international fear. A long time later, vital questions nonetheless stay: How does antimicrobial resistance emerge, and the way can scientists observe the hidden adjustments resulting in it? Why does resistance in some microbes stay undetected till a deadly disease happens? Filling those wisdom gaps is a very powerful to fighting long term outbreaks, bettering remedy results and saving lives.
Antimicrobial resistance may also be fatal.
Over time, my paintings as a microbiologist and biomedical scientist has excited by investigating the genetics of infectious microbes. My colleagues and I known a resistance gene prior to now undetected within the U.S. the usage of genetic and computational strategies that may assist beef up how scientists come across and observe antimicrobial resistance.
Demanding situations of detecting resistance
Antimicrobial resistance is a herbal procedure the place microbes repeatedly evolve as a protection mechanism, obtaining genetic adjustments that reinforce their survival.
Sadly, human actions can accelerate this procedure. The overuse and misuse of antibiotics in well being care, farming and the surroundings push micro organism to genetically trade in ways in which let them live to tell the tale the medicine supposed to kill them.
Early detection of antimicrobial resistance is a very powerful for efficient remedy. Surveillance in most cases starts with a laboratory pattern got from sufferers with suspected infections, which is then analyzed to spot possible antimicrobial resistance. Historically, this has been finished the usage of culture-based strategies that contain exposing microbes to antibiotics within the lab and watching whether or not they survived to decide whether or not they have been turning into resistant. In conjunction with serving to government and researchers track the unfold of antimicrobial resistance, hospitals use this method to come to a decision on remedy plans.
Then again, culture-based approaches have some barriers. Resistant infections frequently pass overlooked till antibiotics fail, making each detection and intervention processes gradual. Moreover, new resistance genes would possibly get away detection altogether.
Genomics of antimicrobial resistance
To triumph over those demanding situations, researchers have built-in genomic sequencing into antimicrobial resistance surveillance. Thru whole-genome sequencing, we will analyze the entire DNA in a microbial pattern to get a complete view of the entire genes provide – together with the ones accountable for resistance. With the computational gear of bioinformatics, researchers can successfully procedure huge quantities of genetic information to beef up the detection of resistance threats.
In spite of its benefits, integrating genomic sequencing into antimicrobial resistance tracking gifts some demanding situations of its personal. Top prices, high quality assurance and a scarcity of skilled bioinformaticians make implementation tough. Moreover, the complexity of deciphering genomic information would possibly prohibit its use in medical and public well being decision-making.
Bioinformatics lets in researchers to research massive organic datasets.
hh5800/iStock by way of Getty Pictures Plus
Setting up global requirements may just assist in making whole-genome sequencing and bioinformatics an absolutely dependable device for resistance surveillance. The International Well being Group recommends laboratories practice strict high quality keep an eye on measures to verify correct and related effects. This comprises the usage of dependable, user-friendly computational gear and shared microbial databases. Further methods come with making an investment in coaching systems and fostering collaborations between hospitals, analysis labs and universities.
Finding a resistance gene
Combining complete genome sequencing and bioinformatics, my colleagues and I analyzed Salmonella samples amassed from a number of animal species between 1982 and 1999. We found out a Salmonella resistance gene known as blaSCO-1 that has avoided detection in U.S. cattle for many years.
The blaSCO-1 gene confers resistance to microbes towards a number of vital antibiotics, together with ampicillin, amoxicillin-clavulanic acid and, to a point, cephalosporins and carbapenems. Those medicines are a very powerful for treating infections in each people and animals.
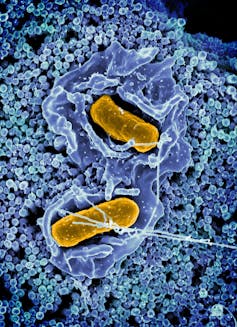
Salmonella Typhimurium invading a mobile.
NIAID/Flickr, CC BY-SA
The blaSCO-1 gene most likely remained unreported as a result of regimen surveillance normally objectives well known resistance genes and it has overlapping purposes with different genes. Gaps in bioinformatics experience will have additionally hindered its identity.
The failure to come across genes like blaSCO-1 raises fear about its possible function in previous remedy disasters. Between 2015 and 2018, the Facilities for Illness Keep watch over and Prevention started enforcing whole-genome sequencing for regimen surveillance of Salmonella. Research performed all over this era discovered that 77% of multistate outbreaks have been connected to cattle harboring resistant Salmonella.
Those neglected genes have vital implications for each meals protection and public well being. Undetected antimicrobial resistance genes can unfold via meals animals, infected meals merchandise, processing environments and agricultural runoff, permitting resistant micro organism to persist and succeed in people. Those resistant micro organism result in infections which are tougher to regard and build up the danger of outbreaks. Additionally, the worldwide motion of other people, cattle and items implies that those resistant lines can simply go borders, turning native outbreaks into international well being threats.
Figuring out new resistance genes now not simplest fills a vital wisdom hole, but it surely additionally demonstrates how genomic and computational approaches can assist come across hidden resistance mechanisms sooner than they pose well-liked threats.
Strengthening surveillance
As antimicrobial resistance continues to upward push, adopting a One Well being way that integrates human, animal and environmental elements can assist be sure that rising resistance does now not outpace people’ skill to battle it.
Tasks just like the Quadripartite AMR Multi-Spouse Believe Fund supply give a boost to for systems that support international collaborative surveillance, advertise accountable antimicrobial use and pressure the advance of sustainable choices. Making sure researchers all over the world practice commonplace analysis requirements will permit extra labs – particularly the ones in low- and middle-income international locations – to give a contribution to international surveillance efforts.
The well being of long term generations will depend on the sector’s skill to verify meals protection and offer protection to public well being on an international scale. Within the ongoing fight between microbial evolution and human innovation, vigilance and flexibility are key to staying forward.




